Research
Our main interest is in the way biologically diverse environments create conditions for macromolecules to associate and dissociate and form complexes that carry specific functions in cells. Examples include receptors that bind or unbind ligands, and pass signals across cell membranes, proteins that fold and unfold in solution, and sometimes form aggregates such as amyloid fibers, or DNA that binds enzymes.
|  |
We have been following this theme in a number of biologically relevant systems that involve collections of macromolecules, such as peptide folding and aggregation, and viral assembly.
|   |
We emphasize theoretical approaches to help dissect the different contributing forces involved, while maintaining close contact to experimental findings. One of the topics we are particularly intereseted in is the sugar trehalose and its effect on proteins. Here's a short talk for a general audience over beer (and Cheeza) --> |
Molecular crowding
Micro-molecular Crowding and Osmotic Effect on Peptide Folding and AggregationNature has developed many strategies to ensure that protein folding occurs in vivo with efficiency and fidelity. Among the most widely employed strategies is the use of small solute molecules called osmolytes that most often confer stability to folded proteins by preferential exclusion from macromolecular surfaces. Recent evidences indicate that modest changes in environmental conditions set by osmolytes and other cosolutes can have profound impacts on protein and peptide conformation and aggregation. One of our main focuses has been to elucidate how proteins and peptides are influenced by the crowded cellular environments. It is hard to imagine that this crowding does not profoundly influence macromolecular interactions in the cellular environment. However, for many years and until recently, this effect has been largely ignored. Our recent efforts to follow this crowding and osmolyte activity yielded surprising results that are changing the way we understand the osmotic action of cosolutes. Specifically, our results contrast the prevailing entropic mechanisms that have been suggested to determine protein stability. Instead, we are seeing that even though solutes are excluded from macromolecular surfaces, the solution and its structure still play dominant roles in determining protein stability. This paradigm shift has also led us to study the formation of amyloid fibers in solution. We find that here, too, osmolytes do not seem to conform to the prevailing ideas of macromolecular crowding. Instead, osmolytes seem to affect amyloid aggregation differently than macromolecular crowders (such as the polymer PEG), in that they delay the time required for fibril nucleation, but increase the amount of fibrils formed at equilibrium. Recent references:
|
|
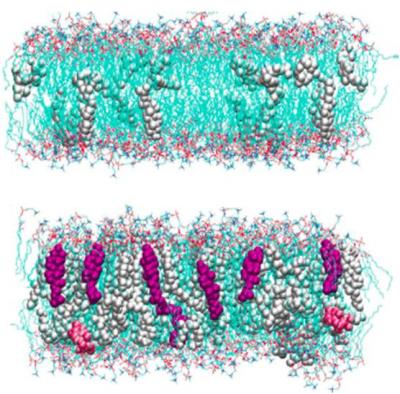
Membrane Interactions
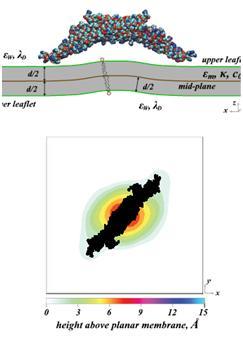
Membrane Interactions & Properties Of Lipid BilayersWe have been probing the interactions of macromolecules at the lipid membrane interface. The material properties of lipids, as well as protein-lipid interactions, govern the interactions and consequent assembly of these multi-component mixtures. The study of such complex systems requires the development of new computational tools that are based on statistical thermodynamics. Membrane Deformations & Lipid Demixing Upon BAR Protein Domain Adsorption
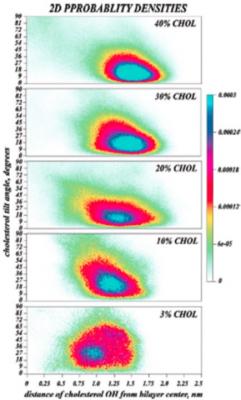
We performed molecular dynamics (MD) simulations of hydrated bilayers containing mixtures of dimyristoylphosphatidylcholine (DMPC) and cholesterol at various ratios, to study the effect of cholesterol concentration on its orientation, and to characterize the link between cholesterol tilt and overall phospholipid membrane organization. The simulations show a substantial probability for cholesterol molecules to transiently orient perpendicular to the bilayer normal, and suggest that cholesterol tilt may be an important factor for inducing membrane ordering. In particular, we find that as cholesterol concentration increases (1-40% cholesterol) the average cholesterol orientation changes in a manner strongly (anti)correlated with the variation in membrane thickness. Furthermore, cholesterol orientation is found to be determined by the aligning force exerted by other cholesterol molecules. To quantify this aligning field, we analyzed cholesterol orientation using, to our knowledge, the first estimates of the cholesterol tilt modulus c from MD simulations. Our calculations suggest that the aligning field that determines is indeed strongly linked to sterol composition. This empirical parameter (c) should therefore become a useful quantitative measure to describe cholesterol interaction with other lipids in bilayers, particularly in various coarse-grained force fields. References:
|
|
Granular Materials
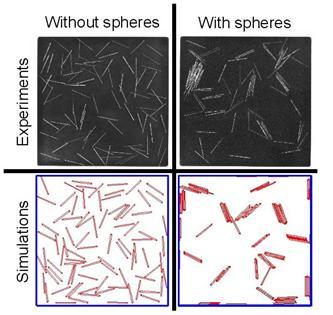
Depletion forces and confinement effects in vibrofluidized granular materials
Ranging from nano- to granular-scales, control of particle assembly can be achieved by limiting the available free space, for example by increasing the concentration of particles (crowding) or through their restriction to 2D environments. It is unclear, however, if self-assembly principles governing thermally equilibrated molecules can also apply to mechanically excited macroscopic particles in nonequilibrium steady-state. We have shown that low densities of vibrofluidized steel rods, when crowded by high densities of spheres and confined to quasi-2D planes, can self-assemble into linear polymer-like structures. Our 2D Monte Carlo simulations show similar finite sized aggregates in thermally equilibrated binary mixtures. Using theory and simulations, we have been able to demonstrate how depletion interactions create oriented ‘‘binding’’ forces between rigid rods to form these ‘‘living polymers.’’ Unlike rod–sphere mixtures in 3D that can demonstrate well-defined equilibrium phases in coexistence, our mixtures confined to 2D lack these transitions because lower dimensionality favors the formation of linear aggregates, thus suppressing a true phase transition. The qualitative and quantitative agreement between equilibrium and granular patterning for these mixtures suggests that entropy maximization is the determining driving force for bundling. Furthermore, this study uncovers a previously unknown patterning behavior at both the granular and nanoscales, and may provide insights into the role of crowding at interfaces in molecular assemblyMoreover, we have found that rods assemble and disassemble to form nematic phases. Interestingly, we have been able to find elastic constants for these systems that would correspond to the appropriate continuum “Frank free energy” that describes molecular nematics. The method relies on analyzing defects in the container to infer the cost of creating such defects in terms of splay and bend contribution to the free-energy. References:
|
|