1. The molecular mechanism underling plant cell competency to regenerate
Certain plants cells retain plasticity and are capable - under the appropriate stimuli, e.g. after wounding or following hormone stimulation - to switch their developmental program, re-enter the cell cycle and form a mass of less-differentiated totipotent cells, termed callus. Those cells restore a “stem cell” status and acquire a competence to respond rapidly to diverse stimuli taking on new fate accordingly and regenerate. The acquisition of new fate is characterized by massive reprogramming of gene expression for example switching on or off hundreds of developmental genes. In eukaryotes, gene expression is the outcome of transcription by RNA polymerase II (Pol II) machinery, acting on the highly dynamic platform of the chromatin and regulated by combinatorial epigenetic modifications. Two of the histone modifications, the active mark H3K4me3, catalyzed by the Trithorax group (TrxG) and the repressive mark H3K27me3 which is catalyzed by the Polycomb Repressive Complex 2 (PRC2) and their interplay can set the gene in an active, repressive or poised for activation transcriptional state.
To elucidate the molecular mechanism underlying plant cell competency to regenerate we are examining the capacity of callus derived from various mutant of the PRC2 and TrxG to response to external cues and to regenerate. Using ChIP-seq analysis we are characterizing the H3k4me3 and H3K27me3 landscapes and mapping the PolII occupation genome wide and relate it to the transcription levels in WT and in the set of mutants. Our goal is to determine the combinatorial code that sets the transcriptional state of each gene into active, repressed or poised for activation in the totipotent callus and to compare it with a non-totipotent leaf cells and the various mutant.
We have identified in callus 800 putative bivalent genes (see below), in which the genes are marked by both H3K27me3 and H3K4me3 and in WT no mRNA. This can serve as a mechanism to set the genes poised for activation. In the emf2 mutant, which is impaired in setting the H3K27me3 mark, those gene Have the H3K4me3 mark and the genes are expressed.
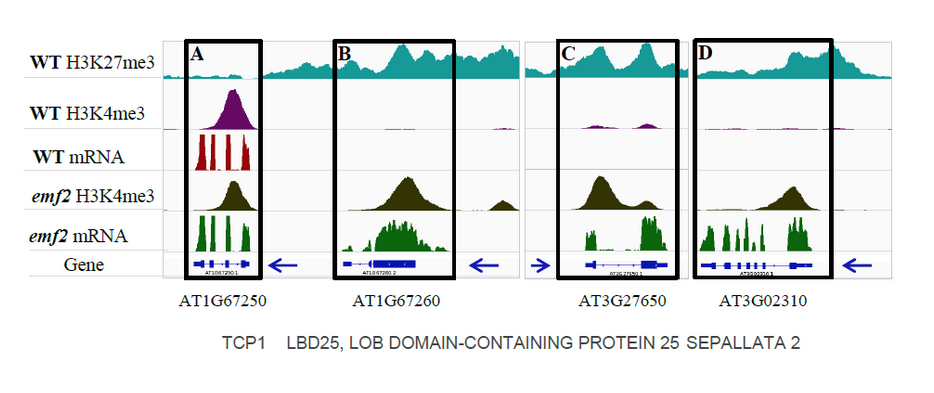
We have identified many other genes that in WT have the H3K27me3 mark and accordingly are not expressed but in the emf2 mutant acquired the H3K4me3 mark and are highly expressed (see below). This mutant does not regenerate properly and by comparing the chromatin landscape we revealed that
| leor_eshed_williams_lab.docx | 429 KB |

