Membrane Interactions & Properties Of Lipid Bilayers
We have been probing the interactions of macromolecules at the lipid membrane interface. The material properties of lipids, as well as protein-lipid interactions, govern the interactions and consequent assembly of these multi-component mixtures. The study of such complex systems requires the development of new computational tools that are based on statistical thermodynamics.
Membrane Deformations & Lipid Demixing Upon BAR Protein Domain Adsorption
Many proteins participating in cellular signaling processes contain BAR domains that have been implicated in membrane shaping. While the BAR domains have been shown to form dimers that are suggested to sense or induce significant curvatures on cell membranes, the underlying mechanisms are not well understood.
To address such processes quantitatively, we have introduced a dynamic mean-field scheme that allows self-consistent calculations of the equilibrium state of membrane-protein complexes after such lateral reorganization of the membrane components, and serves to probe kinetic details of the process. Applicable to membranes with heterogeneous compositions containing several types of lipids, this comprehensive method accounts for mobile salt ions and charged macromolecules in three dimensions, elastic energies of the membrane, as well as for lateral demixing of charged and net-neutral lipids in the membrane plane.
We then applied this new formalism to investigate such mechanisms of protein-membrane interactions, and have studied complexes of single Amphiphysin BAR domains interacting with large patches of lipid membrane bilayers of heterogeneous compositions. Our results suggest that a single BAR dimer is capable of stabilizing a significantly curved membrane, but we predict that such deformations will occur only for membrane patches that have the inherent propensity for high curvature, reflected in the tendency to create local distortions that closely matches the curvature of the BAR dimer itself. Such favorable preconditioning for BAR-membrane interaction may be the result of perturbations such as local lipid demixing induced by the interaction, or of a prior insertion of the BAR domain's amphiphatic N-helix.
Cholesterol Ordering In Lipid Membranes
An essential component of mammalian cell membranes, cholesterol is known to be critical for membrane organization, dynamics, and function. The nonuniform distribution of cholesterol between cellular organelles, lipid membrane compartments, and even between leaflets of the same bilayer, highlights cholesterol’s role in influencing the biophysical properties of a fluid lipid matrix and in the stabilization and function of membrane proteins through specific interactions. The great variability found in the concentration of cholesterol among various cells and between the plasma membrane and the variety of membranes of other cellular organelles underscores the importance of tightly regulated cholesterol content for proper function at the subcellular level. In fact, inborn errors of cholesterol synthesis lead to major developmental abnormalities, and conversely, an excess of cholesterol is widely acknowledged as detrimental.
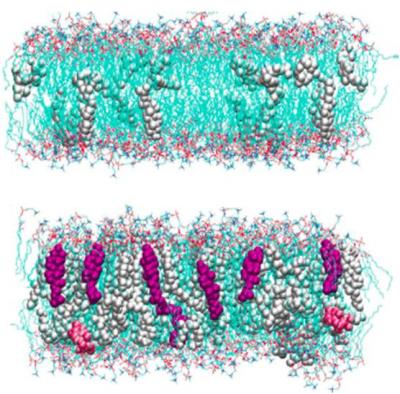
We performed molecular dynamics (MD) simulations of hydrated bilayers containing mixtures of dimyristoylphosphatidylcholine (DMPC) and cholesterol at various ratios, to study the effect of cholesterol concentration on its orientation, and to characterize the link between cholesterol tilt and overall phospholipid membrane organization. The simulations show a substantial probability for cholesterol molecules to transiently orient perpendicular to the bilayer normal, and suggest that cholesterol tilt may be an important factor for inducing membrane ordering. In particular, we find that as cholesterol concentration increases (1-40% cholesterol) the average cholesterol orientation changes in a manner strongly (anti)correlated with the variation in membrane thickness. Furthermore, cholesterol orientation is found to be determined by the aligning force exerted by other cholesterol molecules. To quantify this aligning field, we analyzed cholesterol orientation using, to our knowledge, the first estimates of the cholesterol tilt modulus c from MD simulations. Our calculations suggest that the aligning field that determines is indeed strongly linked to sterol composition. This empirical parameter (c) should therefore become a useful quantitative measure to describe cholesterol interaction with other lipids in bilayers, particularly in various coarse-grained force fields.
References:
- G. Khelashvili, H. Weinstein, D. Harries, Protein Diffusion on Charged Membranes: A Dynamic Mean-Field Model Describes Time Evolution and Lipid Reorganization, Biophys. J. 94, 2580-2597 (2008).
- G. Khelashvili, D. Harries, H. Weinstein, Modeling Membrane Deformations and Lipid Demixing upon Protein-Membrane Interaction: The BAR Dimer Adsorption, Biophys. J. 97, 1626-1635 (2009).
- G. Khelashvili, G. Pabst, D. Harries, Cholesterol Orientation and Tilt Modulus in DMPC bilayers, J. Phys. Chem B 114, 7524-7534 (2010).
- G. Khelashvili, D. Harries, Modeling signaling processes across cellular membranes using a mesoscopic approach. In Ann. Rep. Comp. Chem. Vol. 6, Eds. R.A. Wheeler & N. Baker, 237-261 (2010).
|
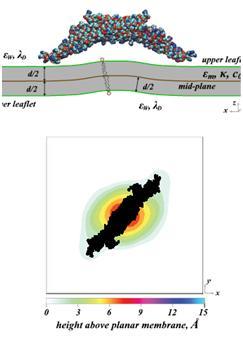
BAR domain adsorbed to lipid membranes causing elastic deformation and lipid segregation, as derived in our mean-field calculations.
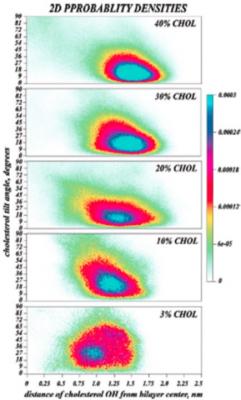
Cholesterol in membranes transition from a broad orientational distribution at low cholesterol mole fractions to a narrow one at high cholesterol fractions. This change in orientation can be seen in the probability distributions as a function of depth in the membrane and tilt angle.
|