Citation:
Abstract:
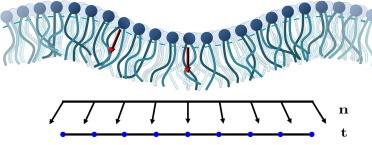
Advances over the past decade have made it possible to extract elastic constants of lipid assemblies from molecular dynamics simulations. We summarize existing strategies for obtaining membrane elastic moduli and clarify the differences in the underlying approaches. We analyze these strategies in depth, including several important advantages and limitations. By addressing these limitations, we obtain a newly formulated spatially local methodology for extracting bending and tilt moduli: The Real Space Instantaneous Surface Method (ReSIS). With its freely available implementation, this method is designed for highly dynamic systems with arbitrary interface geometries. We demonstrate how the method provides consistent results for membranes of arbitrary size. In addition, we describe alternative implementations of various Fourier-space methods, and use these to compare the results from the different available methods and from published computational data. We specifically focus on the tilt modulus, where very large differences between Fourier and real-space based methods are observed, including those derived using ReSIS. These discrepancies are likely due to the known difference between model moduli and thermodynamic moduli that are derived from the corresponding response functions. In addition, we reexamine the issue of angular degeneracy and its effect on conformational ensembles. Finally, a van ’t Hoff analysis of the tilt and bending moduli reveals that both modes act as entropic springs and that enthalpy favors nonzero tilt, perhaps heralding the spontaneous lipid chain tilt of the gel phase at lower temperatures.
Notes:
Additional information and implementation available here